Derda Lab bridges DNA-Encoded Chemistry, Chemical Biology and Machine Learning to solve fundamental problems in chemical reactivity, molecular recognition and drug discovery”
Ongoing Projects
We are interested in developing genetically-encoded ligand discovery and genetically-encoded fragment-based discovery (1) of potent inhibitors with the goal of developing drug candidates or targeting/imaging agents for undraggable targets.
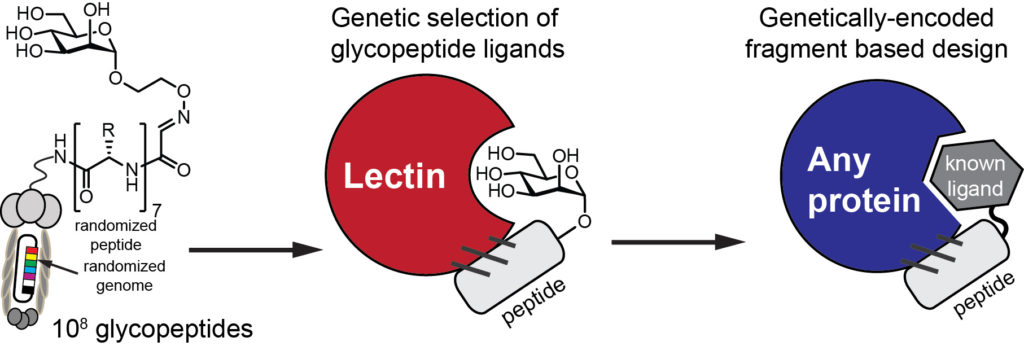
Access to billions of genetically-encoded molecules developed by our group allows solving the “impossible problems in molecular recognition” and identification of ligands for “undruggable targets”: these are molecular receptors for which there are few or no therapeutics available. Proteins that recognize carbohydrates are one of the examples of such target. Protein-carbohydrate interactions are engaged in nearly all physiological responses, such as protein folding and degradation, cell-cell and cell-pathogen interaction, tumor growth and immune responses. Our group pioneered a powerful genetically-encoded discovery of molecules that block or mimic protein-carbohydrate interactions (1,2). To attack these and other unsolved problems in drug discovery, we design approaches that allow building genetically-encoded peptide and macrocyclic of expanded diversity with arbitrary unnatural fragments (3, 4).
1. 4. Ratmir Derda and Simon Ng “Genetically-Encoded Fragment-Based Discovery (GE-FBD)”, Current Opinion in Chemical Biology, 2019, 50, 128.
2. S Ng, MR Jafari, W Matochko, R Derda* “Quantitative Synthesis of Genetically Encoded Glycopeptide Libraries Displayed on M13 Phage” ACS Chem. Biol., 2012, 7 (9), 1482–1487.
3. S. Ng, et al, “Genetically-encoded Fragment-based Discovery of Glycopeptide Ligands for Carbohydrate-binding Proteins“, J. Am. Chem. Soc. 2015, 137, 5248–5251
4. KF Tjhung, S. Ng, PI Kitov, EN Kitova, L Deng, JS Klassen, and R Derda* “Silent Encoding of Chemical Post-Translational Modifications in Phage-Displayed Libraries“, J. Am. Chem. Soc., 2016, 138, 32–35
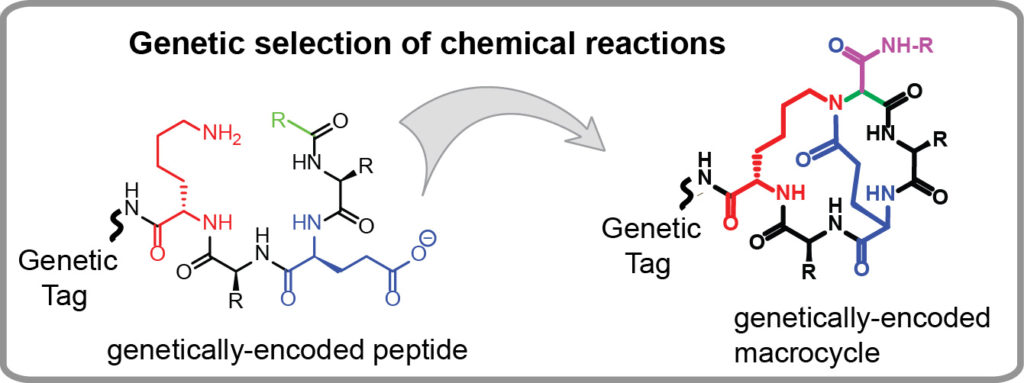
Genetically-encoded libraries of chemicals have emerged as a critical sources of discovery of drug candidates in the pharmaceutical industry. Our group contributes to this field by developing effective approaches for synthesis of genetically encoded libraries by late-stage functionalization. We demonstrated that genetically-encoded libraries of peptides can be successfully modified using a wide range of bio-orthogonal chemical reactions to yield molecules with new function, improved stability or bioavailability. We are interested in
- Identification of new chemical reactions that could modify unprotected peptides, in water, at an ambient temperature and physiological conditions (pH=4-10) to yield products with complex topology (macrocycles, bicycles, etc).
- Employing genetically encoded libraries to study the mechanisms and structure-activity-relationships (SAR) of organic reactions and to discover new reactions.
Our long term goal is to firmly establish new synthesis paradigms in which a chemical space of billion starting materials (e.g., peptides) is converted to a new billion-scale chemical space of products in one simple chemical step. These new chemical spaces will be important starting points to identify valuable precursors to pharmaceuticals, vaccines, diagnostics, and materials. Some examples of synthesis of genetically-encoded libraries by our group are provided below:
1. S. Kalhor-Monfared, M. R. Jafari, J. T. Patterson, P. I. Kitov, J. J. Dwyer, J. J. Nuss and R. Derda “Rapid Biocompatible Macrocyclization of Peptides with Decafluoro-diphenylsulfone” Chem. Sci., 2016, 7, 3785-3790.
2. P Kitov, D F Vinals , S Ng , K F Tjhung , and R Derda “Rapid, Hydrolytically Stable Modification of Aldehyde-terminated Proteins and Phage Libraries”, J. Am. Chem. Soc., 2014, 136, 8149–8152.
3. S. Ng and R Derda* “Phage-displayed macrocyclic glycopeptide libraries” Org. Biomol. Chem., 2016, 14, 5539-5545
4. V. Triana, R. Derda*, “Tandem Wittig / Diels-Alder Diversification of Genetically Encoded Peptide Libraries“, Org. Biomol. Chem., 2017, 15, 7869-7877
5. S Ng, MR Jafari, R Derda* “Bacteriophages and Viruses as a Support for Organic Synthesis and Combinatorial Chemistry“, ACS Chem. Biol. 7, 123.
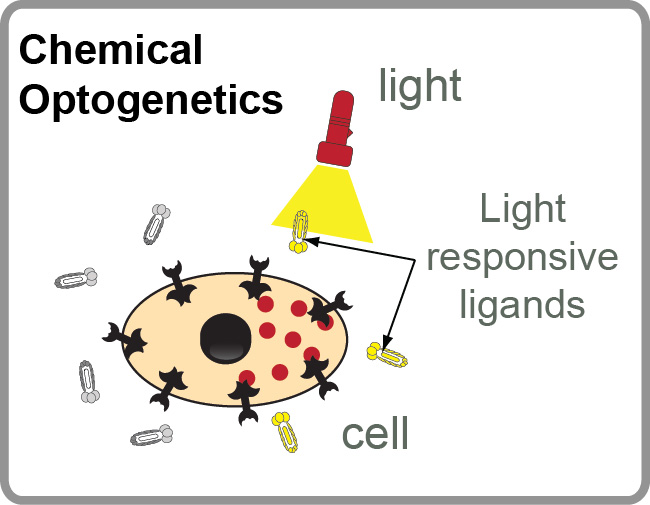
Our Group has long-term interest in “photo pharmacology” and “chemical optogenetics”. The filed of “photopharmacology” aims to develop therapeutics compounds that can be activated by light with spatial and temporal resolution in desired tissue, cell or sub-cellular location. “Chemical optogenetics” is a field on the interface of “chemical genetics” (interrogation of cell function using small-molecule inhibitors) and “optogenetics” (investigation of cell functions using proteins that can be reversibly activated by light). The main challenge in both fields is identification of potent light-responsive small-molecule ligands that agonize or antagonize a desired receptor in the cells. To address this challenge, we are developing methods for genetic selections for ligands that can be reversibly activated or deactivated with light (1-3).
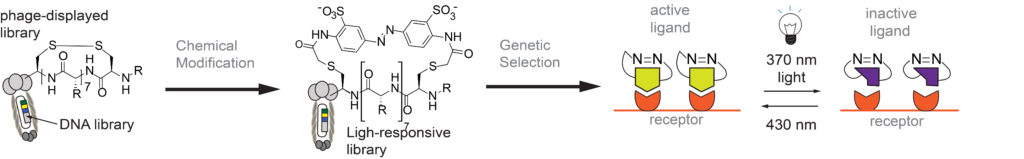
1. MR Jafari, Lu Deng, S Ng, W Matochko, K Tjhung, A Zeberof, A Elias, John S. Klassen, R Derda* “Discovery of light-responsive ligands through screening of light-responsive genetically-encoded library“, ACS Chem. Biol., 2014, 9, 443–450
2. M. R. Jafari, J. Lakusta, R. J. Lundgren, and R Derda* “Allene Functionalized Azobenzene Linker Enables Rapid and Light-Responsive Peptide Macrocyclization” Bioconj. Chem., 2016, 27, 509–514
3. Mohammad R. Jafari, Hongtao Yu, Jessica M. Wickware, Yu-Shan Lin and Ratmir Derda*, “Light Responsive Bicyclic Peptides” Org. Biomol. Chem., 2018,16, 7588-7594.
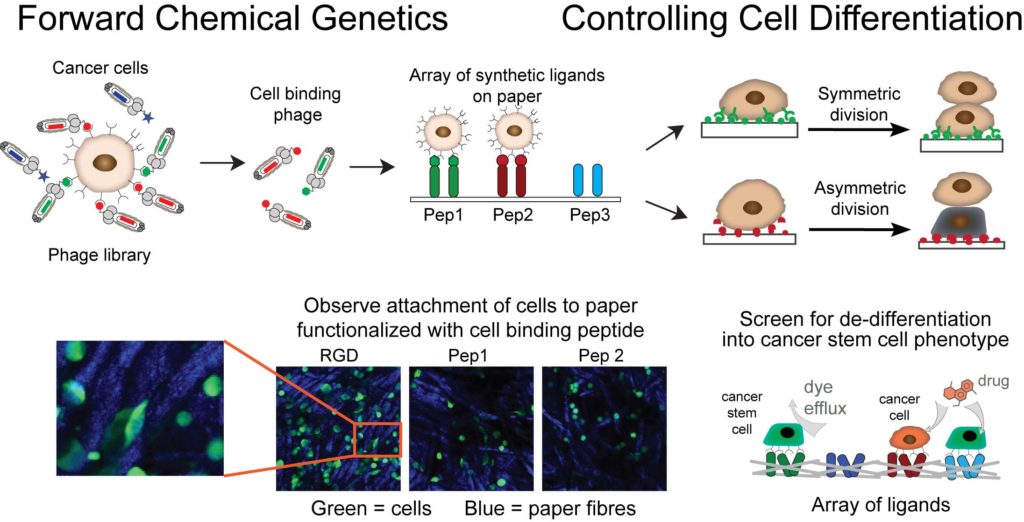
Our goal is to use unsupervised molecular discovery called “forward chemical genetics” to identify molecules and materials that can control differentiation and asymmetric division of cancer stem cells. These events govern tumor development through dynamic control of equilibrium between tumor-initiating cancer stem cells (CSC) and non-stem cancer cells (NSCC). Suppression of the emergence of CSC is critical for halting the growth and relapse of tumors. Large genetically-encoded libraries that contain >109 diverse molecules are uniquely poised for such discovery: by exposing the specific cell type, such as breast CSC, to a mixture of genetically encoded small molecules, we allow the cell to “select” all the molecules that bind to the cellular receptors present on the surface of CSC. This powerful approach requires no knowledge about the structure of composition of molecular receptors and it has the potential to provide all possible ligands for all cellular receptors at once. We couple this unsupervised discovery approach with ligand microarray technology developed by our group to accelerate the discovery of instructive materials that control differentiation of cancer cells to CSC.
1. F Deiss, W L Matochko, N Govindasamy, E Y Lin and R Derda “Flow-Through Synthesis on Teflon-Patterned Paper To Produce Peptide Arrays for Cell-Based Assays”, Angewandte Chemie, 2014, 53, 6374–6377.
2. E. Lin, A. Sikhand, J. Wickware, Y. Hao and R Derda* “Peptide Microarray Patterning for Controlling and Monitoring Cell Growth” Acta Biomater., 2016, 34, 53–59
3. F. Deiss, Y. Yang, W. L. Matochko, R. Derda* “Heat-enhanced peptide synthesis on Teflon-patterned paper“, Org. Biolmol. Chem. 2016, 14, 5148-5156
Vast genetically encoded libraries of chemicals are a powerful source of chemical information with promise to solve many fundamental problems in molecular recognition. However, as any large-scale information source (e.g., Internet, pool of evolutionary traits), search in genetically-encoded libraries requires an efficient “search engine” and fundamental understanding of “discovery trajectories”. Our group aim to understand the fundamental forces that govern these discovery trajectories. We are interested in bridging molecular discovery in genetically encoded libraries with well-established paradigms in physics that successfully deal with information and complexity (Lyapunov stability; Kholmogorov complexity; period doubling bifurcations).
Like internet, vast chemicals space of genetically-encoded libraries contains areas that cannot be easily accessed. Biased molecular discovery can be conceptually compared to search in the same internet using ad-biased search engines vs. searching the internet by bias-free search engines. We develop discovery approach that control the search bias and provide access to outcomes “hidden” to conventional discovery approaches (1). We employ deep-sequencing technology (2), to characterize hundreds of million of DNA sequences at various stages of molecular discovery and we build knowledge clouds that store thousands of such deep-sequencing campaigns and discovery trajectories. As an example, we discovered that distribution of molecular libraries to micro-droplets removes the unwanted biases in the selection landscape by isolating library members from one another and ceasing the unwanted competition between them (3,4). Genetically-encoded screens confined to compartments made by emulsion yield ligands significantly different from those found in conventional screens.
1. Derda R, Tang SKY, Li SC, Ng S, Matochko WL, Jafari MR. “Diversity of Phage-Displayed Libraries of Peptides During Panning and Amplification” Molecules 2011, 16, 1776-1803
2. W Matochko, K Chu, B Jin, SW Lee, G Whitesides, R Derda* “Deep sequencing analysis of phage libraries using Illumina“, Methods, 2012, 58 (1), 47–55
3. (a) Derda R, Tang SKY, Whitesides GM “Uniform Amplification of Phage with Different Growth Characteristics in Monodisperse Droplet-Based Compartments” Angew. Chem. Intl. Ed. 2010 49(31), 5301–5304; (b) Matochko, S Ng, MR Jafari, J Romaniuk, SKY Tang, R Derda* “Uniform amplification of phage display libraries in monodisperse emulsions“, Methods, 2012, 58 (1), 18-27; (c) K. F. Tjhung, S. Burnham, H. Anany, M. W. Griffiths and R. Derda “Rapid enumeration of phage in monodisperse emulsions“, Anal. Chem., 2014, 86, 5642–5648.
4. W Matochko, SC Li, SKY Tang, R Derda * “Prospective identification of parasite sequences in phage display screens” Nuc. Acid. Res., 2014, 42, (3), 1784-1798.
The Central Dogma of Biology, DNA → RNA → Protein, facilitates the study of Biology using a unified toolbox of DNA sequencing and DNA association approaches . This ability has revolutionized and transformed all areas of biomedical and life science. Carbohydrates are one of the most diverse and ubiquitous components in all kingdoms of life but investigating the biological roles of carbohydrates cannot rely on DNA sequencing directly. We developed technology that that introduces the missing one-to-one correspondence between DNA sequence and carbohydrate structure [1]. Genetically-encoded multivalent Liquid Glycan Array (LiGA) [1-3] and Liquid Lectin Array (LiLA) [4] enable application of powerful deep sequencing approaches to rapidly accumulate the critical information about interactions of glycans and glycan binding proteins in vitro and in vivo.
Information provided by technology like LiGA and similar technologies can be combined with machine learning approaches to solve the fundamental long-standing problem of “glycan code”: given a sequence of a glycan and sequence (or structure) of protein, predict how strongly glycan and protein interact with one another [5].
1. Mirat Sojitra, Susmita Sarkar, Jasmine Maghera, Emily Rodrigues, Eric J. Carpenter, Shaurya Seth, Daniel Ferrer Vinals, Nicholas J. Bennett, Revathi Reddy, Amira Khalil, Xiaochao Xue, Michael R. Bell, Ruixiang Blake Zheng, Ping Zhang, Corwin Nycholat, Justin J. Bailey, Chang-Chun Ling, Todd L. Lowary, James C. Paulson, Matthew S. Macauley & Ratmir Derda* “Genetically encoded multivalent liquid glycan array displayed on M13 bacteriophage“, Nature Chemical Biology 2021, 17, 806–816
2. Chih-Lan Lin, Mirat Sojitra, Eric J. Carpenter, Ellen S. Hayhoe, Susmita Sarkar, Elizabeth A. Volker, Chao Wang, Duong T. Bui, Loretta Yang, John S. Klassen, Peng Wu, Matthew S. Macauley, Todd L. Lowary & Ratmir Derda “Chemoenzymatic synthesis of genetically-encoded multivalent liquid N-glycan arrays” Nature Comm. 14, 5237 (2023)
3. M.Sojitra, E. N. Schmidt, G. M. Lima, E. J. Carpenter, K. A. McCord, A. Atrazhev, M. S. Macauley, R. Derda, “Measuring carbohydrate recognition profile of lectins on live cells using liquid glycan array (LiGA)” Nature Protocols (2024)
4. G. M. Lima, Z. Jame-Chenarboo, M. Sojitra, S. Sarkar, E. J. Carpenter, C. Y. Yang, E. Schmidt, J. Lai, A. Atrazhev, D. Yazdan, C. Peng, E. A. Volker, R. Ho, G. Monteiro, R. Lai, L. K. Mahal, M. S. Macauley, R. Derda, “The liquid lectin array detects compositional glycocalyx differences using multivalent DNA-encoded lectins on phage“, Cell Chem. Biol. (2024)
5. Eric J. Carpenter, Shaurya Seth, Noel Yue, Russell Greiner, and Ratmir Derda* “GlyNet: A Multi-Task Neural Network for Predicting Protein-Glycan Interactions“, Chem. Sci., 2022,13, 6669-6686