2026 Publications
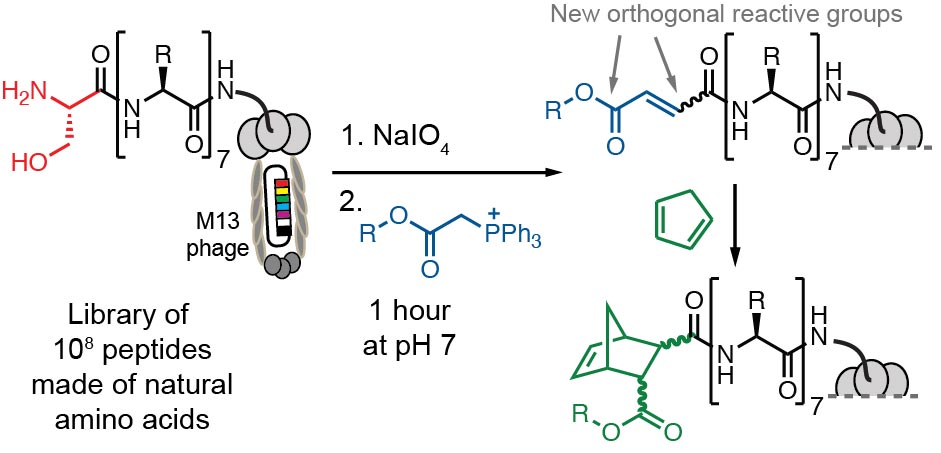
95. J. H. Walker, K. Yan, R. Derda, “From Transient to Stable: Incorporating Electrophiles in Genetically-Encoded and DNA-Encoded Libraries of Peptide-Derived Macrocycles” Biochem. 2026, DOI: 10.1021/acs.biochem.5c00646
2025 Publications
94. E. J. Carpenter, C. Peng, S. Haregu, N. Twells, L. Woudstra, A. Sood, J. Cartmell, R. J. Woods, L. K. Mahal, S.- K. Wang, R. Greiner, R. Derda, “Atom-level machine learning of protein-glycan interactions and cross-chiral recognition in glycobiology” Sci. Adv. 2025, DOI: 10.1126/sciadv.adx6373
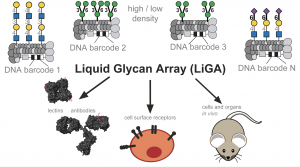
93. M. Sojitra, E. N. Schmidt, G. M. Lima, E. J. Carpenter, K. A. McCord, A. Atrazhev, M. S. Macauley, R, Derda, “Measuring carbohydrate recognition profile of lectins on live cells using liquid glycan array (LiGA)” Nat. Protoc. 20, 989-1019 (2025). DOI: 10.1038/s41596-024-01070-3
92. ,
91. , , , ,
2024 Publications
90. K. Yan, M. Miskolzie, F. Banales Mejia, C. Peng, A. I. Ekanayake, D. J. Maly, R. Derda, “Late-Stage Reshaping of Phage-Displayed Libraries to Macrocyclic and Bicyclic Landscapes using a Multipurpose Linchpin” J. Am. Chem. Soc. 2024, DOI: 10.1021/jacs.4c13561
89. G. M. Lima, Z. Jame-Chenarboo, M. Sojitra, S. Sarkar, E. J. Carpenter, C. Y. Yang, E. Schmidt, J. Lai, A. Atrazhev, D. Yazdan, C. Peng, E. A. Volker, R. Ho, G. Monteiro, R. Lai, L. K. Mahal, M. S. Macauley, R. Derda, “The liquid lectin array detects compositional glycocalyx differences using multivalent DNA-encoded lectins on phage“, Cell Chem. Biol. (2024)
88. M.Sojitra, E. N. Schmidt, G. M. Lima, E. J. Carpenter, K. A. McCord, A. Atrazhev, M. S. Macauley, R. Derda, “Measuring carbohydrate recognition profile of lectins on live cells using liquid glycan array (LiGA)” Nat Protoc. (2024)
87. K. Yan, M. Miskolzie, F. Banales Mejia, C. Peng, A. I. Ekanayake, D. J. Maly, R. Derda, “Late-stage reshaping of phage-displayed libraries to macrocyclic and bicyclic landscapes using multipurpose linchpin” ChemRxiv 2024, DOI 10.26434/chemrxiv-2024-08847-v2
2023 Publications
86. J.Y.K. Wong, S. E. Kirberger, R. Qiu, A. I. Ekanayake, P. Kelich, S. Sarkar, E. R. Alvizo-Paez, J. Miao, S. Kalhor-Monfared, J. J. Dwyer, J. M. Nuss, Y.-S. Lin, M. S. Macauley, L. Vukovic, W. C.K. Pomerantz, R. Derda, “Genetically-Encoded Discovery of Perfluoroaryl-Macrocycles that Bind to Albumin and Exhibit Extended Circulation in-vivo ” Nat. Comm. 14, 5654 (2023)
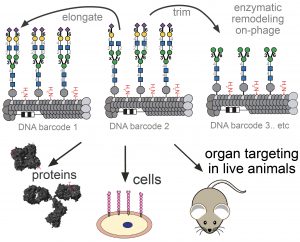
85. Chih-Lan Lin, Mirat Sojitra, Eric J. Carpenter, Ellen S. Hayhoe, Susmita Sarkar, Elizabeth A. Volker, Chao Wang, Duong T. Bui, Loretta Yang, John S. Klassen, Peng Wu, Matthew S. Macauley, Todd L. Lowary & Ratmir Derda “Chemoenzymatic synthesis of genetically-encoded multivalent liquid N-glycan arrays” Nat. Comm. 14, 5237 (2023)
84. K. M. Koss, T. Son, C. Li, Y. Hao, J. Cao, M. A. Churchward, Z. J. Zhang, J. A. Wertheim, R. Derda, K. G. Todd. “Towards discovering a novel family of peptides targeting neuroinflammatory states of brain microglia and astrocytes”, J. Neurochem., 2023, doi doi.org/10.1111/jnc.15840
83. E. N. Schmidt, D. Lamprinaki, K. A. McCord, M. Joe, M. Sojitra, A. Waldow, J. Nguyen, J. Monyror, E. N. Kitova, F. Mozaneh, X. Y. Guo, J. Jung, J. R. Enterina, G. C. Daskhan, L. Han, A. R. Krysler, C. R. Cromwell, B. P. Hubbard, L. J. West, M. Kulka, S. Sipione, J. S. Klassen, R. Derda, T. L. Lowary, M. S. Macauley “Siglec-6 mediates the uptake of extracellular vesicles through a noncanonical glycolipid binding pocket” Nat. Comm. 14: 2327 (2023)
82. M. G. Alteen, R. W. Meek, S. Kolappan, J. A. Busmann, J. Cao, Z. O’Gara, R. Derda, G. J. Davies, D. J. Vocadlo “Phage display uncovers a sequence motif that drives polypeptide binding to a conserved regulatory exosite of O-GlcNAc transferase” Proc. Natl. Acad. Sci. USA, 120 (42) e2303690120 (2023)

2022 Publications
81. J.Y.K. Wong, S. E. Kirberger, R. Qiu, A. I. Ekanayake, P. Kelich, S. Sarkar, E. R. Alvizo-Paez, J. Miao, S. Kalhor-Monfared, J. J. Dwyer, J. M. Nuss, Y.-S. Lin, M. S. Macauley, L. Vukovic, W. C.K. Pomerantz, R. Derda, Genetically-Encoded Discovery of Perfluoroaryl-Macrocycles that Bind to Albumin and Exhibit Extended Circulation in-vivo, doi: https://doi.org/10.1101/2022.08.22.504611, BioRxiv 2022 (accepted to Nat. Comm, see pub #86 above)
80. C.-L. Lin, M. Sojitra, E. J. Carpenter, E. S. Hayhoe, S. Sarkar, E. A. Volker, A. Atrazhev, Todd L. Lowary, M. S. Macauley, R. Derda “Chemoenzymatic Synthesis of Genetically-Encoded Multivalent Liquid N-glycan Arrays“, BioRxiv 2022, DOI https://doi.org/10.1101/2022.08.05.503005 (accepted to Nat. Comm, see pub #85 above)
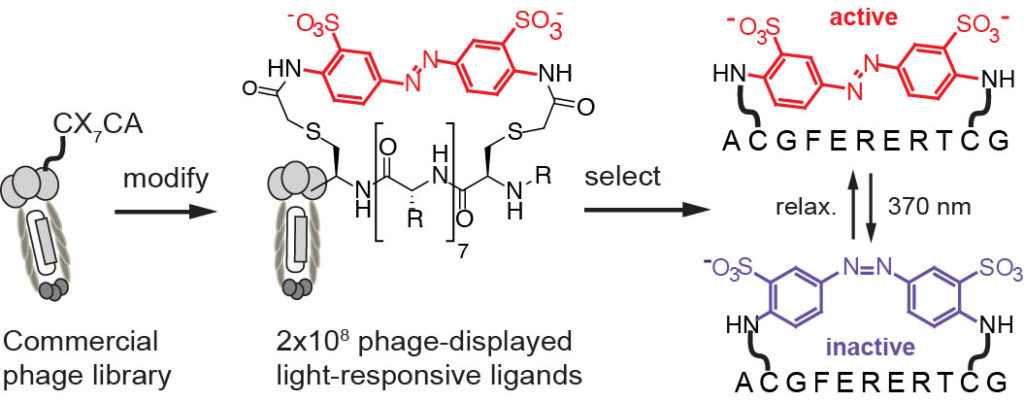
79. Lena Sobze, Ratmir Derda, “Development and Characterization of Light-Responsive Peptide Macrocycles“. Methods Mol Biol. 2022, 2371:411-426.
78. Natalia Milosevich, Katrina H Vizely, Roko PA Nikolic, Jacob F McCallum, Lee M Treanor, Sarah S Khan, Ratmir Derda, Fraser Hof, “Installation of cysteine-derived methyllysine mimics on phage-dis-played peptide libraries: optimization of reaction conditions for conversion and phage viability“, ChemRxiv 2022, DOI 10.26434/chemrxiv-2022-1lkkx
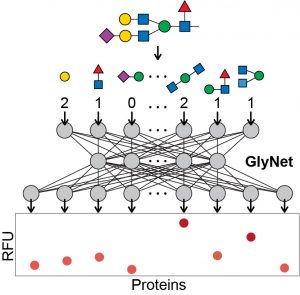
77. Eric J. Carpenter, Shaurya Seth, Noel Yue, Russell Greiner, and Ratmir Derda* “GlyNet: A Multi-Task Neural Network for Predicting Protein-Glycan Interactions“, Chem. Sci., 2022,13, 6669-6686 and BioRxiv Preprint 2021, YouTube videos: OneGlycan1246Proteins, OneProtein599Glycans, OneProtein4160NewGlycans
2021 Publications
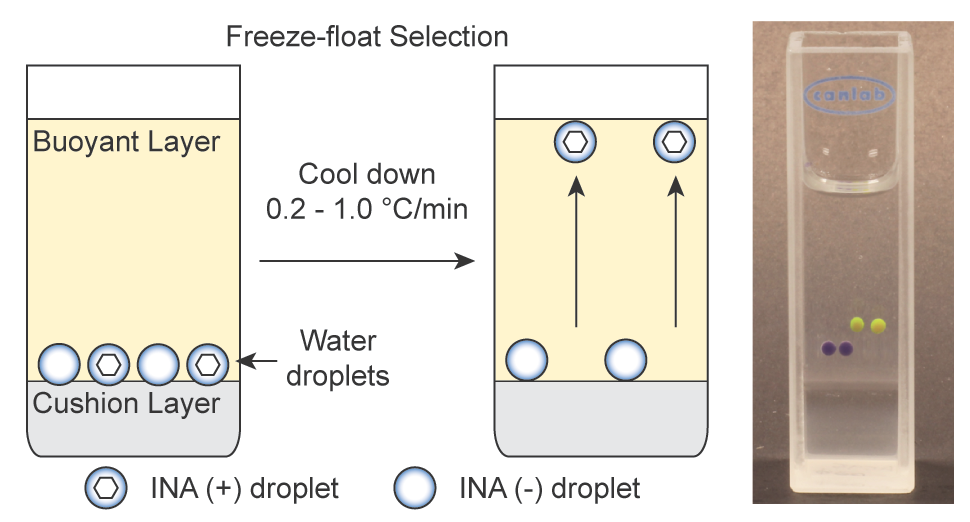
76. Yuki Kamijo Ratmir Derda, “Freeze-Float System for High-throughput Measurement of Ice Nucleation“, ChemRXiv, DOI10.26434/chemrxiv-2021-19l62 2021
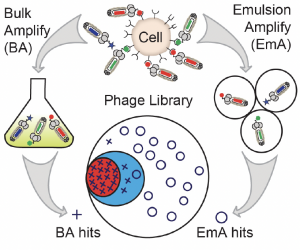
75. W. L. Matochko, F. Deiss, Y. Yang, R. Derda* “Reproducible Discovery of Cell-Binding Peptides ‘Lost’ in Bulk Amplification via Emulsion Amplification in Phage Display Panning“. BioRvix Preprint 2021, DOI 2021.10.31.466683v1
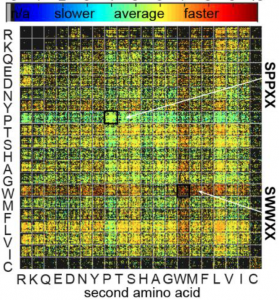
74. Kejia Yan, Vivian Triana, Sunil Vasu Kalmady, Kwami Aku-Dominguez, Sharyar Memon, Alex Brown, Russell Greiner, Ratmir Derda* “Learning Structure Activity Relationship (SAR) of the Wittig Reaction from Genetically-Encoded Substrates“, Chem. Sci., 2021,12, 14301-14308. ChemRxiv Preprint 2021 DOI https://doi.org/10.26434/chemrxiv.14609670.v1 (48HD.cloud link)
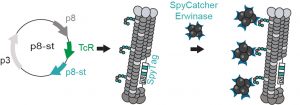
73. Guilherme M. Lima, Alexey Atrazhev, Susmita Sarkar, Mirat Sojitra, Revathi Reddy, Matthew S. Macauley, Gisele Monteiro*, and Ratmir Derda* “DNA-Encoded Multivalent Display of Protein Tetramers on Phage: Synthesis and In Vivo Applications” BioRxiv. Preprint 2021 doi: https://doi.org/10.1101/2021.02.20.432100 (48HD.cloud link) ACS Chemical Biology 17 (11), 3024-3035, 2021.
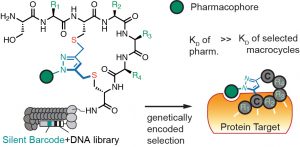
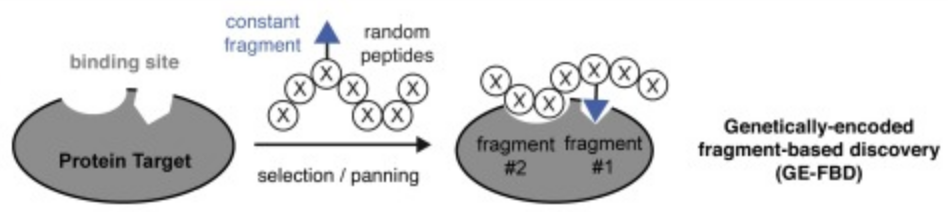
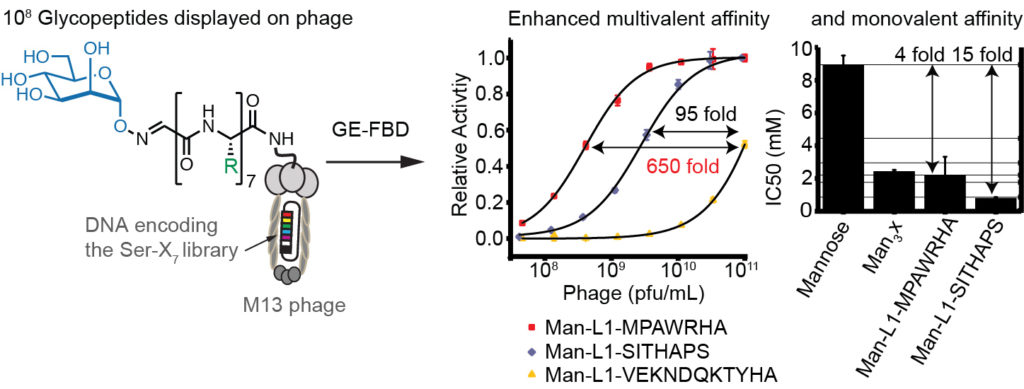
72. Arunika I. Ekanayake, Lena Sobze, Payam Kelich, Jihea Youk, Nicholas J. Bennett, Raja Mukherjee, Atul Bhardwaj, Frank Wuest, Lela Vukovic, Ratmir Derda , “Genetically Encoded Fragment-Based Discovery (GE-FBD) from Phage-Displayed Macrocyclic Libraries with Genetically-Encoded Unnatural Pharmacophores” J. Am. Chem. Soc. 2021 143, 14, 5497–5507 (48HD.cloud link). Highlight: Christian Heinis “Combining biological and chemical diversity“. Nature Chemistry 2021, 13 (6) , 512-513.
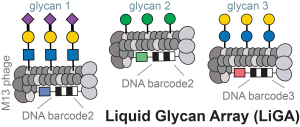
71. Mirat Sojitra, Susmita Sarkar, Jasmine Maghera, Emily Rodrigues, Eric J. Carpenter, Shaurya Seth, Daniel Ferrer Vinals, Nicholas J. Bennett, Revathi Reddy, Amira Khalil, Xiaochao Xue, Michael R. Bell, Ruixiang Blake Zheng, Ping Zhang, Corwin Nycholat, Justin J. Bailey, Chang-Chun Ling, Todd L. Lowary, James C. Paulson, Matthew S. Macauley & Ratmir Derda* “Genetically encoded multivalent liquid glycan array displayed on M13 bacteriophage“, Nature Chemical Biology 2021, 17, 806–816.
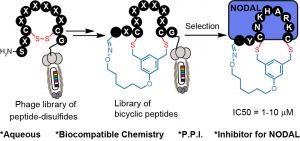
70. Jeffrey Wong, Raja Mukherjee, Olena Bilyk, Jiayuan Miao, Vivian Triana Guzman, Mark Miskoizie, Yu-Shan Lin, Antoine Henninot, John J. Dwyer, Serhii Kharchenko, Anna Iampolska, Dmitriy Volochnyuk, Lynne M. Postovit, Ratmir Derda* “Genetically-Encoded Discovery of Proteolytically Stable Bicyclic Inhibitors of Morphogen NODAL“, Chem. Sci., 2021,12, 9694 (48HD.cloud link) ChemRxiv Preprint 2021 DOI https://doi.org/10.26434/chemrxiv.13870592.v1
2020 Publications
69. Arunika Ekanayake, Lena Sobze, Jihea Youk, Nicholas J. Bennett, Raja Mukherjee, Atul Bhardwaj, Frank Wuest, Ratmir Derda, “Genetically Encoded Fragment-Based Discovery (GE-FBD) from Phage-Displayed Macrocyclic Libraries with Genetically-Encoded Unnatural Pharmacophores“ ChemRxiv. Preprint., 2020 DOI: 10.26434/chemrxiv.12644225.v1 See the final publication in J. Am. Chem. Soc. 2021 143, 14, 5497–5507. Article Twitter Feed: @RatmirDerda, @Derdalab
68. M. Sojitra, S. Sarkar, J. Maghera, E. Rodrigues, E. J. Carpenter, S. Seth, D. F. Vinals, N. J. Bennett, R. Reddy, A. Khalil, X. Xue, M. Bell, R. B. Zheng, P. Zhang, CC Ling, T. L. Lowary, J. C. Paulson, M. S. Macauley, R. Derda* “Genetically Encoded, Multivalent Liquid Glycan Array (LiGA)“, BioRXiv, 2020, DOI: 10.1101/2020.03.24.997536 SI available at BioRXiv and LigaCloud.ca: highlighted on Faculty Opinion. Article Twitter Feed: @RatmirDerda @derda_lab, @MITglycobio, @CarolynBertozzi, @ChemicalBiology
2019 Publications
67. Elena N Kitova, Ling Han, Daniel F Vinals, Pavel I Kitov, Ratmir Derda, John S Klassen*, “Influence of labeling on the glycan affinities and specificities of glycan-binding proteins. A case study involving a C-terminal fragment of human galectin-3“, Glycobiology, 2019, 30, 49-57.
66. Ratmir Derda* and Simon Ng “Genetically-Encoded Fragment-Based Discovery (GE-FBD)”, Current Opinion in Chemical Biology, 2019, 50, 128.
65. Yuki Kamijo and Ratmir Derda*“Freeze-Float Selection of Ice Nucleators” Langmuir, 2019, 35 (2), 359–364
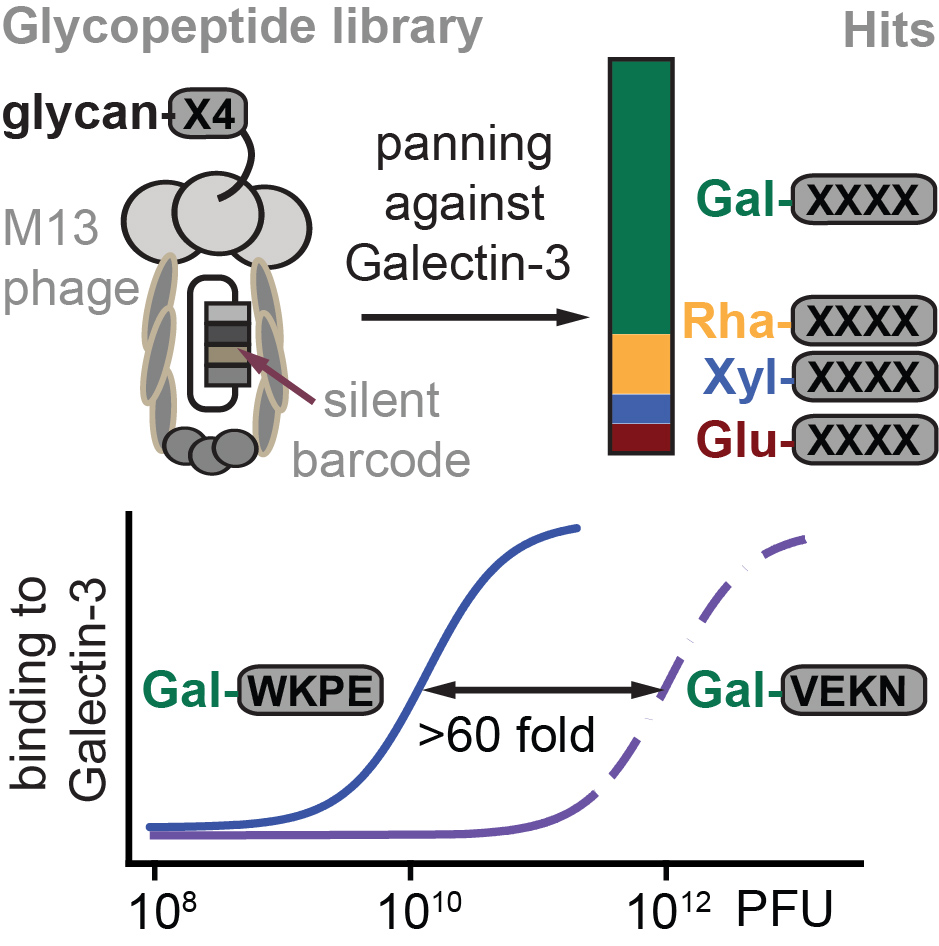
64. Daniel Ferrer Vinals, Pavel I. Kitov, Zhijay Tu, Chunxia Zou, Christopher W. Cairo, Hans Chun-Hung Lin, Ratmir Derda* “Selection of galectin-3 ligands derived from genetically encoded glycopeptide libraries“, Peptide Sci., 2019, 111, e24097 (48HD.cloud link)
63. Bifang He, AM Dzisoo, Ratmir Derda, Jian Huang*. “Development and Application of Computational Methods in Phage Display Technology”, Curr. Med. Chem., 2019, 26, 7672.
62. L Ning, B He, P Zhou, R Derda, J Huang, Molecular Design of Peptide-Fc fusion Drugs, Curr. Drug Metabolism, 2019, 20(3), 203-208.
2018 Publications
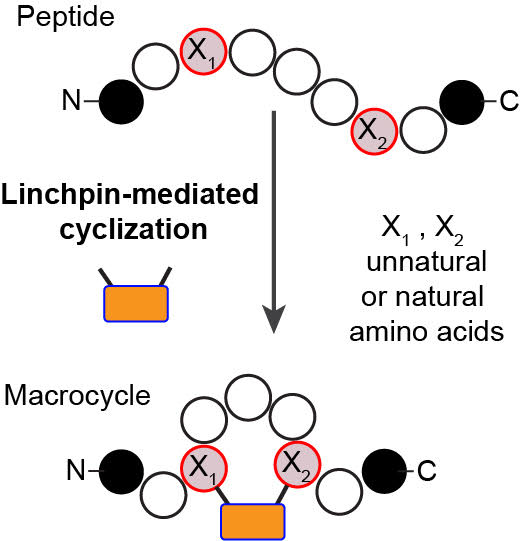
61. Ratmir Derda and Mohammad R Jafari, “Synthetic Cross-linking of Peptides: Molecular Linchpins for Peptide Cyclization”, Prot. Pep. Lett., 2018, DOI : 10.2174/0929866525666181120090650
60. Simon Ng, Nicholas James Bennett, Jessica Schulze, Nan Gao, Christoph Rademacher, Ratmir Derda* “Genetically-encoded Fragment-based Discovery of Glycopeptide Ligands for DC-SIGN“, Bioorg. & Med. Chem., 2018, 26(19), 5368-5377. (48HD.cloud link)
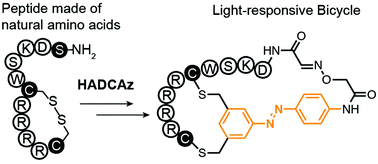
59. Mohammad R. Jafari, Hongtao Yu, Jessica M. Wickware, Yu-Shan Lin and Ratmir Derda*, “Light Responsive Bicyclic Peptides” Org. Biomol. Chem., 2018,16, 7588-7594. Highlighted on RSC blog: “Creating a platform for the development of photoswitchable ligand libraries”

58. Bifang He, Lixu Jiang, Yaocong Duan, Guoshi Chai, Yewei Fang, Juanjuan Kang, Min Yu, Ning Li, Zhongjie Tang, Pengcheng Yao, Pengcheng Wu, Ratmir Derda, Jian Huang, “Biopanning data bank 2018: hugging next generation phage display “, Database, 2018, bay032. DOI: 10.1093/database/bay032
57. B. He, K. F. Tjhung, N. J. Bennett, Y. Chou, A. Rau, J. Huang, R. Derda*, “Compositional Bias in Naïve and Chemically-modified Phage-Displayed Libraries uncovered by Paired-end Deep Sequencing” Nat. Sci. Reports., 2018, 8, 1214 (48HD.cloud link)
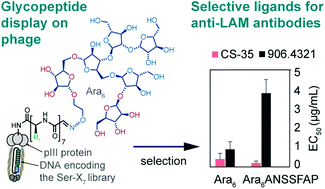
56. Y. Chou, E. N. Kitova , M. Joe, R. Brunton, T. L. Lowary, J. S. Klassen, R. Derda*, “Genetically-encoded fragment-based discovery (GE-FBD) of glycopeptide ligands with differential selectivity for antibodies related to mycobacterial infections“, Org. Biomol. Chem, 2018, 16, 223 – 227 (48HD.cloud link)
2017 Publications
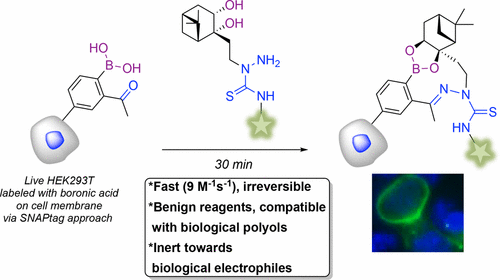
55. B. Akgun, C. Li, Y. Hao, G. Lambkin, R. Derda, D. Hall*, “Synergic ‘Click’ Boronate/Thiosemicarbazone System for Fast and Irreversible Bioorthogonal Conjugation in Live Cells” J. Am. Chem. Soc., 2017, 139, 14285–14291
54. V. Triana, R. Derda*, “Tandem Wittig / Diels-Alder Diversification of Genetically Encoded Peptide Libraries“, Org. Biomol. Chem., 2017, 15, 7869-7877
53. A Adini, I Adini, Z Chi, R Derda, AE Birsner, BD Matthews, RJ D’Amato*, “A novel strategy to enhance angiogenesis in vivo using the small VEGF-binding peptide PR1P“, Angiogenesis, 2017, 20, 399–408.
51. H. Anany, Y. Chou, S. Cucic, S. Evoy, R. Derda, MW Griffiths*, “From Bits and Pieces to Whole Phage to Nanomachines: Pathogen Detection Using Bacteriophage“, Annu. Rev. Food Sci. Tech., 2017, 8, 305-329.
2016 Publications
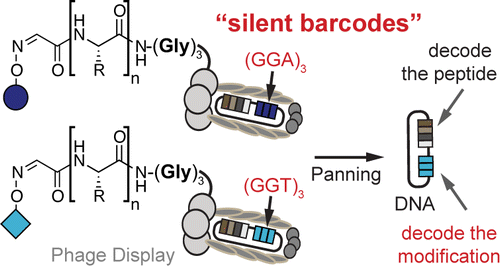
51. K. F. Tjhung, S. Ng, P. I. Kitov, E. N. Kitova, L. Deng, J. S. Klassen, and R. Derda* “Silent Encoding of Chemical Post-Translational Modifications in Phage-Displayed Libraries“, J. Am. Chem. Soc., 2016, 138, 32–35.
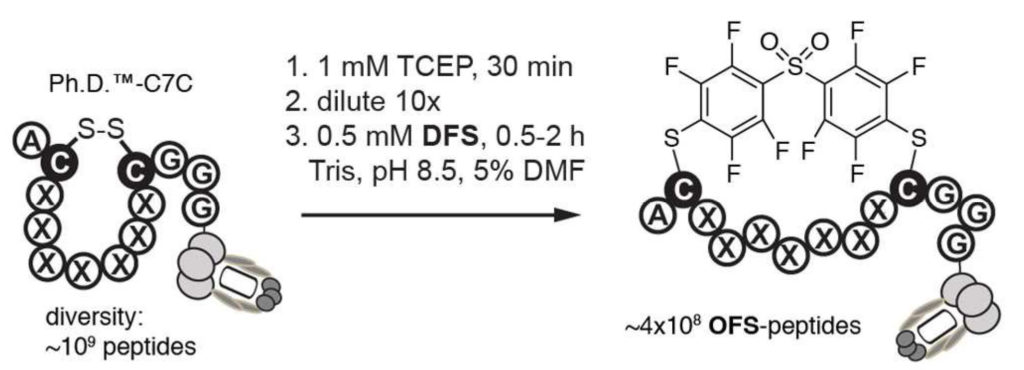
50. S. Kalhor-Monfared, M. R. Jafari, J. T. Patterson, P. I. Kitov, J. J. Dwyer, J. J. Nuss and R. Derda “Rapid Biocompatible Macrocyclization of Peptides with Decafluoro-diphenylsulfone” Chem. Sci., 2016, 7, 3785-3790.
49. S. Ng and R Derda* “Phage-displayed Macrocyclic Glycopeptide Libraries” Org. Biomol. Chem., 2016, 14, 5539-5545
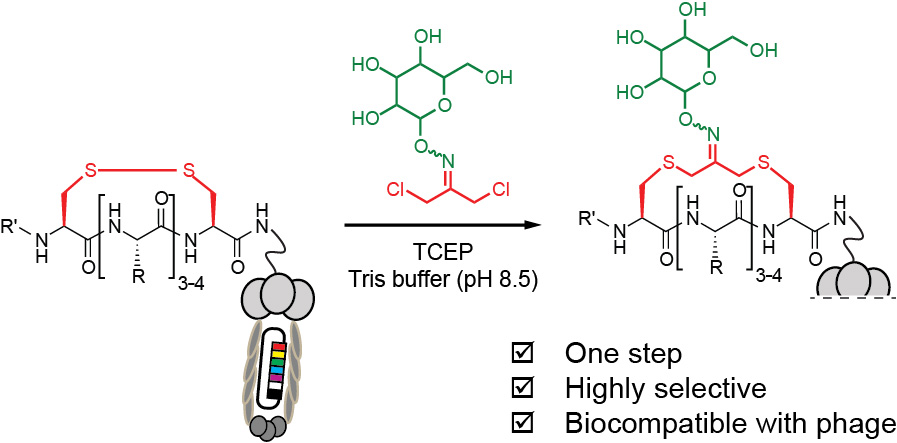
48. F. Deiss, Y. Yang, R. Derda* Parallel Syntheses of Peptides on Teflon-Patterned Paper Arrays (SyntArrays). (book chapter) Methods. Mol. Biol. 2016, 1368, 249-71
47. M. R. Jafari, J. Lakusta, R. J. Lundgren, and R Derda* “Allene Functionalized Azobenzene Linker Enables Rapid and Light-Responsive Peptide Macrocyclization” Bioconj. Chem., 2016, 27, 509–514
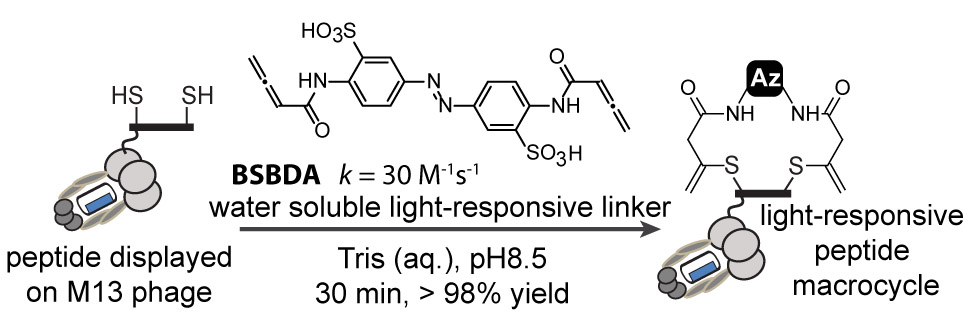
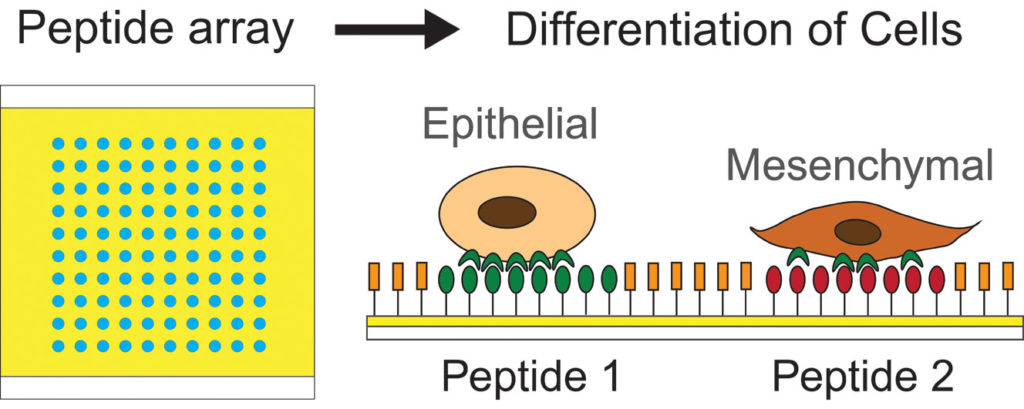
46. E. Lin, A. Sikhand, J. Wickware, Y. Hao and R Derda* “Peptide Microarray Patterning for Controlling and Monitoring Cell Growth” Acta Biomater., 2016, 34, 53–59 download PDF
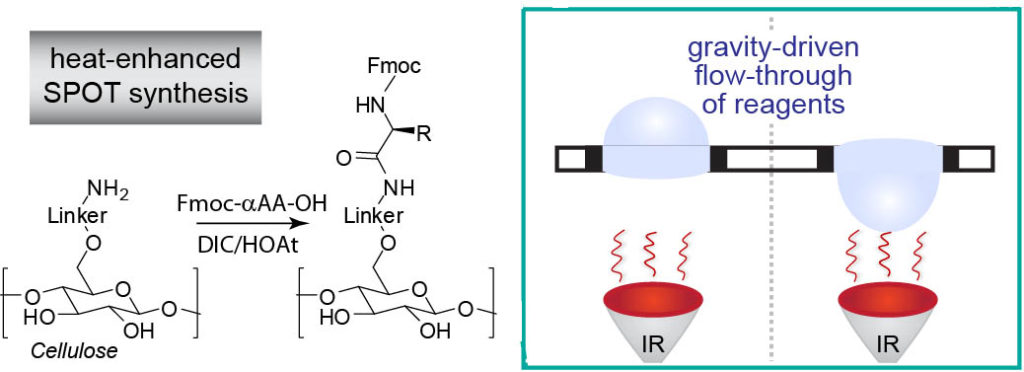
45. F. Deiss,# Y. Yang,# W. L. Matochko, R. Derda* “Heat-enhanced peptide synthesis on Teflon-patterned paper“, Org. Biolmol. Chem. 2016, 14, 5148-5156 Download PDF
2015 Publications
44. S. Ng, E. Y. Lin, PI Kitov, KF Tjhung, OO Gerlits, L Deng, B Kasper, A Sood, BM Paschal, P Zhang, CC Ling, JS Klassen, CJ Noren, LK Mahal, RJ Woods, L Coates, and R Derda* “Genetically-encoded Fragment-based Discovery of Glycopeptide Ligands for Carbohydrate-binding Proteins“, J. Am. Chem. Soc., 2015, 137, 5248–5251. Highlight in “Carbohydrates: Inhibitors in an Instant” Nature Chemical Biology 2015, 11, 381, and Practical Fragments: Fragments Ontology
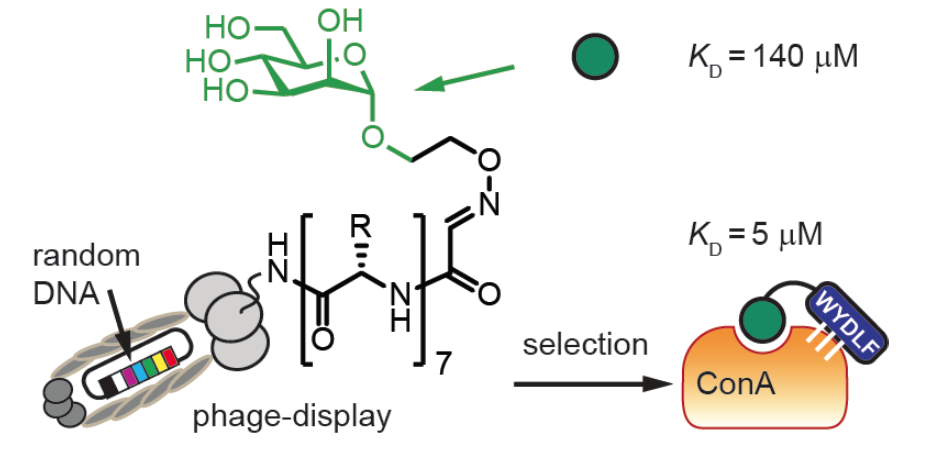
43. K. F. Tjhung, F. Deiss, J. Tran, Y. Chou, R. Derda*, “Intra-domain phage display (ID-PhD) of peptides and mini-domain proteins censored from canonical pIII phage display“, 2015 Front. Microbiol. 6:340.
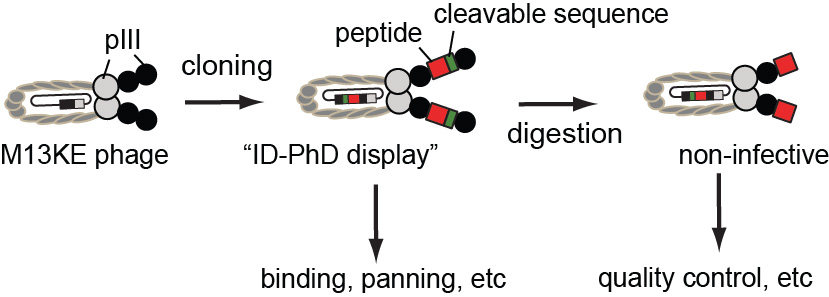
42. Ratmir Derda*, Jesse Gitaka, Thomas M. Kariuki, Catherine M. Klapperich, Charles R. Mace, Ashok A. Kumar, Marya Lieberman, Jacqueline Linnes, Joerg Jores, Johnson Nasimolo, Joseph Ndung’u, Evans Taracha, Abigail Weaver, Douglas B. Weibel, and Paul Yager “Enabling the development and deployment of next generation point of care diagnostics“, 2015, PLoS Negl Trop Dis 9(5): e0003676
41. (Book chapter) W. L. Matochko, R. Derda*, “Next-Generation Sequencing of Phage-Displayed Peptide Libraries“, Methods in Molecular Biology, Volume 1248, 2015, pp 249-266
40. (Book chapter) S. Ng, K. F., Tjhung, B. M. Paschal, C. J. Noren, R. Derda*, “Chemical Posttranslational Modification of Phage-Displayed Peptides“, Methods in Molecular Biology, Volume 1248, 2015, pp 155-172
39. (Book) Ratmir Derda (Editor): Peptide Libraries: Methods and Protocols (Springer), 2015, Vol. 1248. ISBN 978-1-4939-2020-4. Preface: Peptide Libraries on the Chemistry-Biology Interface. This Book is dedicated to the memory of Carlos Barbas the IIIrd

2014 Publications
38. M. Pan, L. Rosenfeld, M. Kim, M. Xu, E. Y. Lin, R. Derda and S. K. Y. Tang “Fluorinated Pickering Emulsions Impede Interfacial Transport and Form Rigid Interface for the Growth of Anchorage-Dependent Cells” ACS Appl. Mater. Interfaces, 2014, 6, 21446–21453
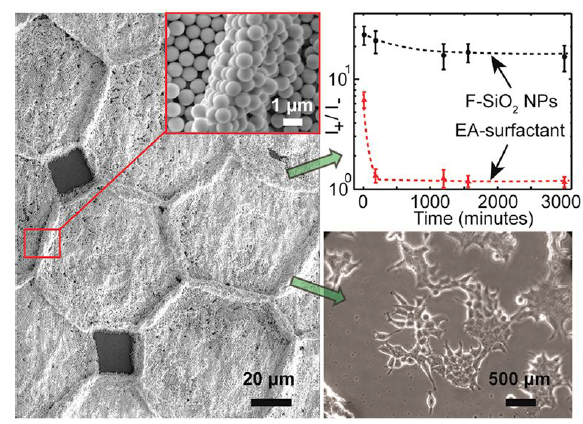
37. S. Burnham, J. Hu, H. Anany, L. Brovko, F. Deiss, R. Derda, M. W. Griffiths*, “Towards rapid on-site phage-mediated detection of generic Escherichia coli in water using luminescent and visual readout“. Anal Bioanal Chem., 2014, 406, (23), 5685-5693
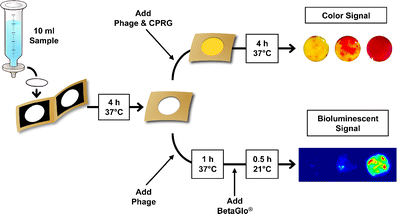
36. P Kitov, D F Vinals , S Ng , K F Tjhung , and R Derda “Rapid, Hydrolytically Stable Modification of Aldehyde-terminated Proteins and Phage Libraries”, J. Am. Chem. Soc., 2014, 136, 8149–8152.
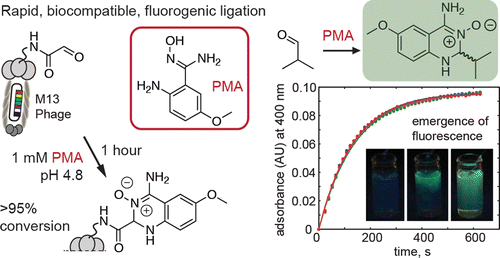
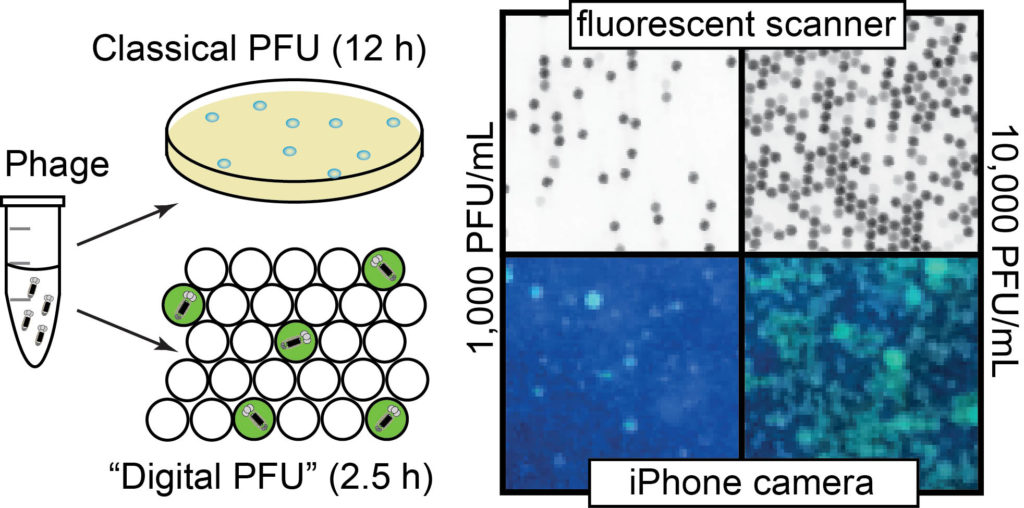
35. K. F. Tjhung, S. Burnham, H. Anany, M. W. Griffiths and R. Derda “Rapid enumeration of phage in monodisperse emulsions“, Anal. Chem., 2014, 86, 5642–5648.
34. F Deiss, W L Matochko, N Govindasamy, E Y Lin and R Derda “Flow-Through Synthesis on Teflon-Patterned Paper To Produce Peptide Arrays for Cell-Based Assays”, Angewandte Chemie, 2014, 53, 6374–6377. (featured as ‘Hot Article’ and on back cover) Download PDF
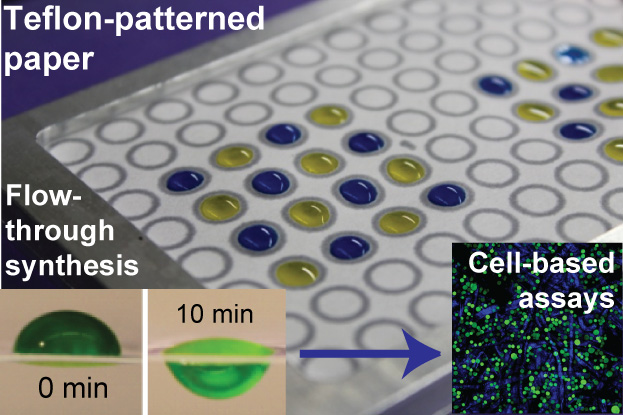
Highlighted in Chemical and Engineering News : “Peptide Synthesis: Nonstick barriers help build arrays for screening”
33. T Lin, Y Chen, L Rosenfeld, R Derda, SKY Tang “Review and Analysis of Performance Metrics of Microfluidic Droplet Systems for Diagnostics and High Throughput Screening Applications” Microfluidics and NanoFluidics, 2014, 16, (5), 921-939
32. MR Jafari, Lu Deng, S Ng, W Matochko, K Tjhung, A Zeberof, A Elias, John S. Klassen, R Derda* “Discovery of light-responsive ligands through screening of light-responsive genetically-encoded library“, ACS Chem. Biol., 2014, 9, 443–450 (front cover)
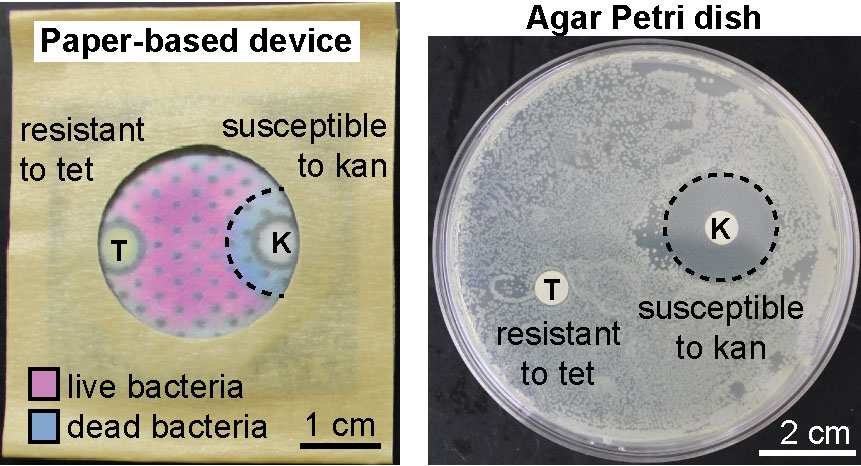
31. F Deiss, MF Huacca, J Bal, K Tjhung, R Derda “Antimicrobial susceptibility assays in paper-based portable culture devices“, Lab Chip, 2014, 14, 167-171, (back cover, and featured as HOT article). See our article highlighted in Chemistry World, on CHEMIE.de information portals (bionity.com, analytica-world.com), and in here
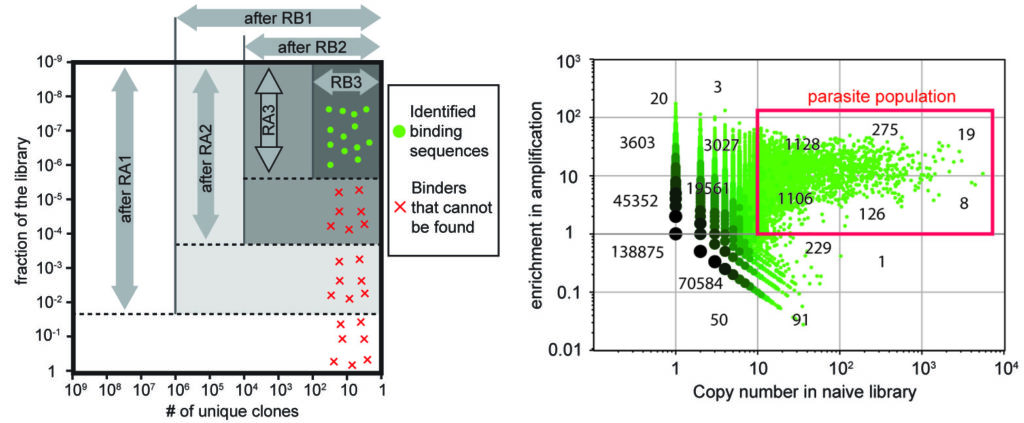
30. W Matochko, SC Li, SKY Tang, R Derda * “Prospective identification of parasite sequences in phage display screens” Nuc. Acid. Res., 2014, 42, (3), 1784-1798.
data pages: illumina processing scripts, raw data files
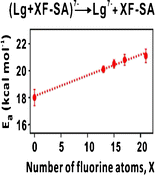
29. L Liu, N Jalili, A Baergen, S Ng, J Bailey, R Derda, JS Klassen* “Fluorine Bonding Enhances the Energetics of Protein-Lipid Binding in the Gas Phase“, Journal of The American Society for Mass Spectrometry, 2014, 25, (5), 751-757
28. BM Scott, WL Matochko, RF Gierczak, V Bhakta, R Derda, WP Sheffield* “Phage display of the serpin alpha-1 proteinase inhibitor randomized at consecutive residues in the reactive centre loop and biopanned with or without thrombin“, PLoS One, 2014, 9, (1), e84491
2013 Publications
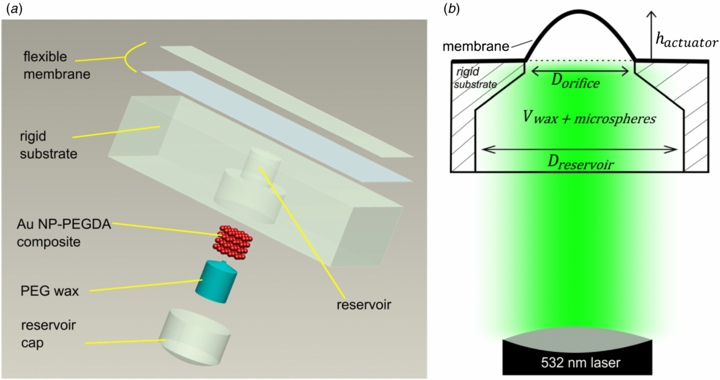
26. A Zeberoff, R Derda, AL. Elias* “Remote activation of a microactuator using a frequency specific photo-responsive gold nanoparticle composite“, J. Micromech. Miroeng., 2013, 23, 125022
25. W Matochko, R Derda “Error analysis of deep sequencing of phage libraries: Peptides censored in sequencing” Comput Math Methods Med. (Phage Display Bioinformatics Issue), 2013, Vol. 2013, Article ID 491612
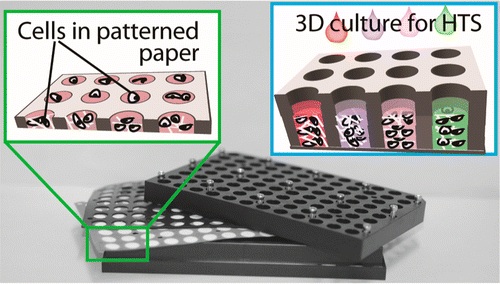
24. F Deiss, A Mazzeo, E Hong, DE Ingber, R Derda,* GM Whitesides* “A Platform for High-Throughput Testing of the Effect of Soluble Compounds on 3D Cell Cultures” Anal. Chem., 2013, 85, 8085–8094
2012 Publications
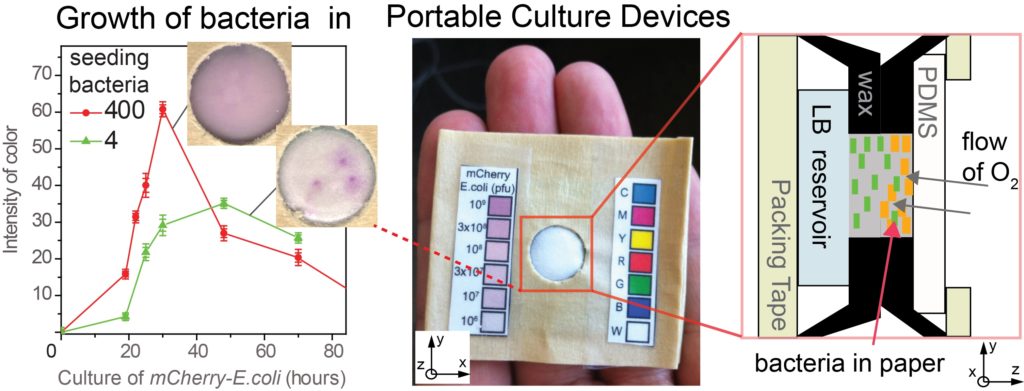
23. MF Huacca, A Wu, E Szepesvari, P Rajendran, N Kwan-Wong, A Razgulin, Y Shen, J Kagira, R Campbell, R Derda* “Portable cultures for phage and bacteria made of paper and tape” Lab Chip, 2012, 12, 4269-4278. Lab on a Chip Top 10% article
22. S Ng, MR Jafari, W Matochko, R Derda* “Quantitative Synthesis of Genetically Encoded Glycopeptide Libraries Displayed on M13 Phage” ACS Chem. Biol., 2012, 7 (9), 1482–1487. Cover Story of September Issue
21. W Matochko, K Chu, B Jin, SW Lee, G Whitesides, R Derda* “Deep sequencing analysis of phage libraries using Illumina“, Methods, 2012, 58 (1), 47–55
20. W Matochko, S Ng, MR Jafari, J Romaniuk, SKY Tang, R Derda* “Uniform amplification of phage display libraries in monodisperse emulsions“, Methods, 2012, 58 (1), 18-27.
19. S Ng, MR Jafari, R Derda* “Bacteriophages and Viruses as a Support for Organic Synthesis and Combinatorial Chemistry“, ACS Chem. Biol., 2012, 7, 123.
18. Derda R, Bracher PJ “YouTube or You Lose: Grand Challenges Canada Explores Whether Scientists Are Ready for Web-Based Grant Competitions” ACS Chem. Biol,20116(8), 771-774
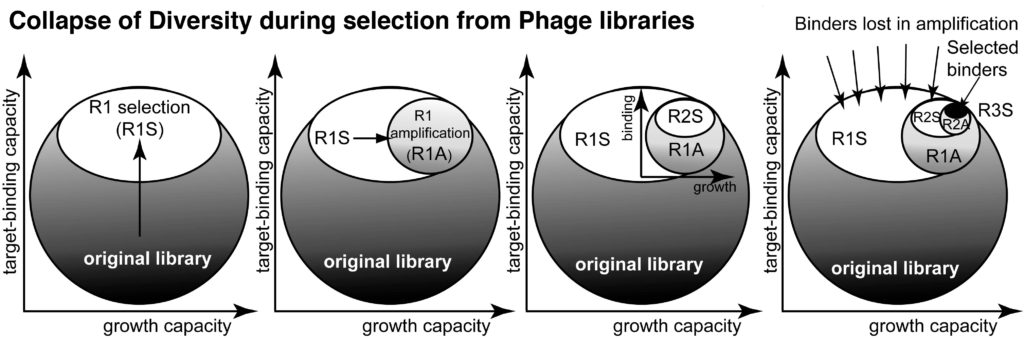
17. Derda R, Tang SKY, Li SC, Ng S, Matochko WL, Jafari MR. “Diversity of Phage-Displayed Libraries of Peptides During Panning and Amplification” Molecules 2011, 16, 1776-1803
Graduate and Postdoctoral Work
16. Gulden Camci-Unal, Anna Laromaine, Estrella Hong, Ratmir Derda, and George M. Whitesides*, “Biomineralization Guided by Paper Templates“, Sci. Rep., 2016, 6016; 6: 27693.
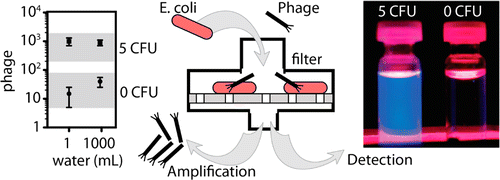
15. R Derda, MR. Lockett, SKY Tang, RC Fuller, EJ Maxwell, B Breiten, CA Cuddemi, A Ozdogan, GM Whitesides “Filter-Based Assay for Escherichia coli in Aqueous Samples Using Bacteriophage-Based Amplification” Anal. Chem., 2013, 85 , 7213–7220
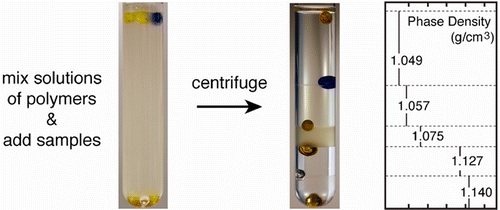
14. C Mace, O Akbulut, A Kuma, N Shapiro, R Derda, M Patton, GM Whitesides* “Aqueous Multiphase Systems of Polymers and Surfactants Provide Self-Assembling Step-Gradients in Density” J. Am. Chem. Soc., 2012, 134, pp 9094–9097
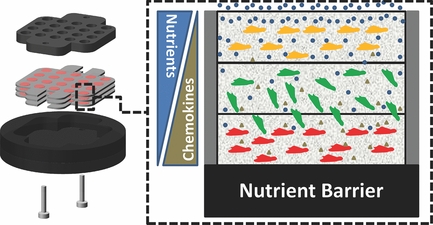
13. B. Mosadegh, B. E. Dabiri, M. R. Lockett, R. Derda, P. Campbell, K. K. Parker, G. M. Whitesides “Three-Dimensional Paper-Based Model for Cardiac Ischemia“, Adv. Healthcare Mater., 2014, 3, 1036–1043.
12. Tang SKY, Derda R, Quan Q, Loncar M, Whitesides GM “Continuously Tunable Droplet-based Optical Microcavities” Optics Express 2011, 19, 2204. This article was selected for publication in the Virtual Journal for Biomedical Optics, Volume 6, Issue 2, 2011.
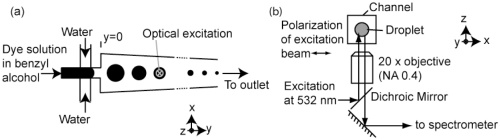
11. Derda R, Tang SKY, Laromaine A, Mosadegh B, Hong E, Mwangi M, Mammoto A, Ingber DE, Whitesides GM “Multizone paper platform for 3D cell cultures“ PLoS One2011, 6, e18940. Highlighted in Biotechniques: “Will paper cell cultures catch fire?”
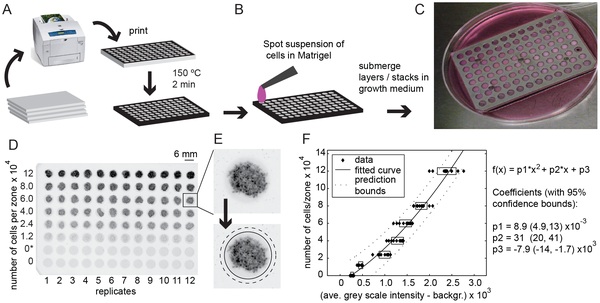
10. Tang SKY, Derda R, Mazzeo AD, Whitesides GM, “Reconfigurable Self-Assembly of Mesoscale Optical Components at a Liquid-Liquid Interface”, Adv. Mater., 2011, 23, 2413.

9. Li L, Klim JR, Derda R, Courtney AH, Kiessling LL, “Spatial control of cell fate using synthetic surfaces to potentiate TGF-beta signaling” Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 11745. Highlighted in C&E News 2011, 89, 6.
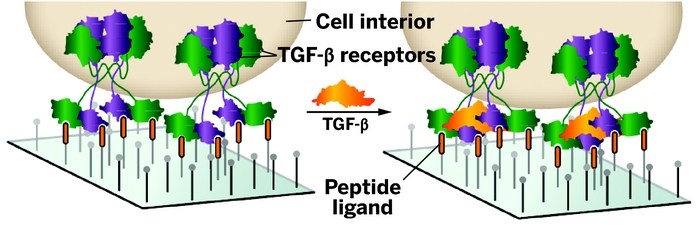
8. Mammoto T, Mammoto A, Tat T, Gibbs A, Derda R, Mannix R, Bruijn M, Yung CW, Huh D, Ingber DE, “Mechanochemical Control of Mesenchymal Condensation and Embryonic Tooth Organ Formation” Dev. Cell 2011, 21, 758
7. Derda R, Tang SKY, Whitesides GM “Uniform Amplification of Phage with Different Growth Characteristics in Individual Compartments Consisting of Monodisperse Droplets” Angew. Chem. Intl. Ed. 2010 49(31), 5301–5304. Highlighted in Faculty of 1000 Biology
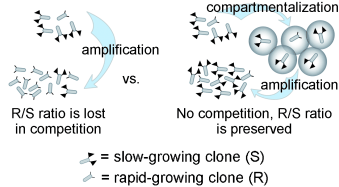
6. Derda R, Musah S, Orner BP, Klim JR, Li L, Kiessling LL “High-throughput Discovery of Synthetic Surfaces that Support Proliferation of Pluripotent Cells” J. Am. Chem. Soc. 2010, 132, 1289. Highlighted in Faculty of 1000 Biology
5. Norville JE, Derda R, Gupta S, Drinkwater KA, Belcher AM, Leschziner AE, Knight TFJr. “Introduction of customized inserts for streamlined assembly and optimization of BioBrick synthetic genetic circuits” J. Biol. Eng. 2010, 4, 17.
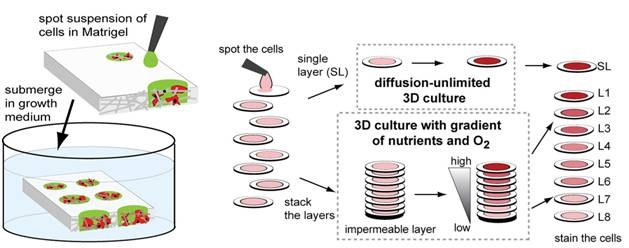
4. Derda R, Laromaine A, Mammoto A, Tang SKY, Mammoto T, Donald Ingber, Whitesides GM “Paper-Supported Three-Dimensional Cell Culture for Tissue-Based Bioassays” Proc. Natl. Acad. Sci. 2009, 106, 18457. Highlighted in Faculty of 1000 Biology, Faculty of 1000 Medicine and Nature Methods 2009, 6, 865.
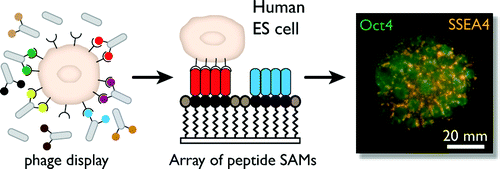
3. Derda R, Ligyin Li, Orner BP, Lewis RL, Thomson JA, Kiessling LL “Defined Substrates for Human Embryonic Stem Cell Growth Identified from Surface Arrays”ACS Chem. Biol. 2007, 2, 347
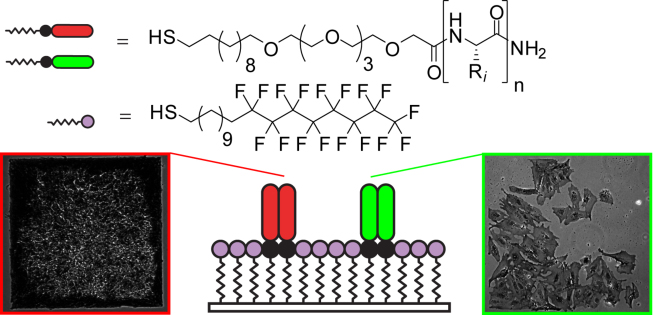
2. Derda R, Wherritt DJ, Kiessling LL “Solid-phase Synthesis of Alkanethiols for the Preparation of Self-Assembled Monolayers on Gold” Langmuir 2007, 23, 11164.
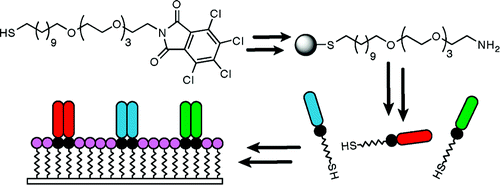
1. Orner BP, Derda R, Lewis RL, Thomson JA, Kiessling LL “The Design and Fabrication of Arrays for the Combinatorial Exploration of Cell Adherence” J. Am. Chem. Soc. 2004, 126, 10808-10809; The article was highlighted in Chemical and Engineering News 2004, 82, 21.